Electrochemistry
Liquid chromatographic electrochemical detection of peptides using the biuret reaction
The biuret reaction between Cu(II) and a peptide. The complexes are electroactive: a one-electron oxidation to Cu(III) occurs at modest potentials for tetrapeptides and longer but is accessible even for dipeptides.
We use the reaction of Cu(II) with peptides in basic solutions to derivatize peptides following their separation by HPLC (reversed phase).
As the scheme shows, the amide protons dissociate, and stable complexes with three nitrogen and one oxygen donor (NNNO) or four nitrogen donors (NNNN) are formed. As Dale Margerum showed in a series of papers in the '70s and '80s, these complexes are electroactive. We have extended those studies, having worked on over 30 bioactive peptides from dipeptides to insulin (Chen 1996; Woltman 1995; Chen 1995)
The postcolumn reaction involves the addition of Cu(II) in basic copper tartrate after the separation. Particularly with microcolumn separations, it is important to avoid adding bandspreading. As we have shown, we can mix effluent and reagents effectively (Woltman et al., 1999, 2000; Sahlin 2002).
Single-cell electroporation
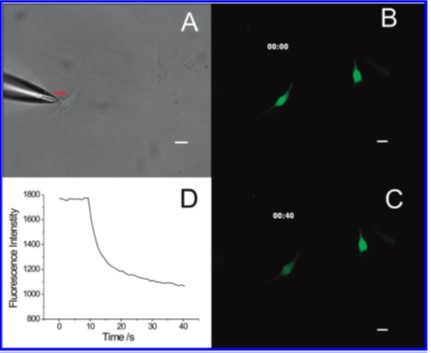
A. Pipette addressing a single A549 cell. B. Before electroporation. Fluorescence is from thiol, chiefly glutathione, labeling with a Thioglo reagent. C. 40 s after. D. Fluorescence vs time demonstrating electroporation and resealing of the membrane.
Single cell electroporation (Nolkrantz 2001, Wang 2010) is a versatile technique for creating transiently permeable cell membranes. We began in collaboration with Owe Orwar, then at Chalmers University of Technology, to determine the electric field in solution near a carbon fiber microelectrode (Lambie 2009). It turns out to be simpler to use a micropipette as the source of the electric field (Nolkrantz 2001). This can be used to transfect single cells, for example (Wang 2009). We have been interested in using this technique to acquire samples of the intracellular contents without sacrificing the cell. Thus, considerable effort has gone to optimizing the conditions for electroportation. Guided by theory (Zudans 2007) on the transmembrane potential during electroporation and how it varies with experimental conditions, we can find experimentally conditions that give a significant probability of successfully electroporating a cell and having that cell survive (Agarwal 2007a, 2007b, 2009).
Electroosmosis in brain tissue
What is the best way to pass fluid through a tissue, or perfuse the tissue, reproducibly and without bias in order to investigate metabolic, biochemical, and other processes? Using pressure could work, but pressure-induced flow velocity is dramatically influenced by the size and geometry of the fluidic pathways in the extracellular space. On the other hand, electroosmosis less affected. Would electroosmosis work in brain tissue? We determined that it does (Guy 2008, Guy 2009). The zeta potential is in the range of 20-25 mV which is quite substantial. We have completely characterized the process of using electroosmosis to drive fluid through brain tissue (Guy 2012), specifically organotypic hippocampal slice cultures, and now use it routinely for the determination of ectopeptidase activity.
Electroosmosis in organotypic hippocampal cultures. A nearly neutral, fluorescently labeled dextran is introduced into the membrane below the tissue (upper and lower spots) and into the culture (large central spot). A voltage is applied with the polarity shown in the left panel. Over time, central and right panels, the dextran in the tissue culture moves with the electroosmotic flow.